Design
What is the BGB structure?
In this project, we introduce the ‘BGB structure’ as a new concept for multilayered liposomal vesicles. BGB structures are three-layered vesicles composed of Giant Unilamellar Vesicles (GUVs), and DNA hydrogel. The three layers consist of an inner lipid membrane, DNA hydrogel, and outer lipid membrane(Fig.1.a). The acronym ‘BGB’ is derived from the initials of each biomolecule in sequence from the inner layer. When DNA strands are used instead of DNA hydrogel, it is termed the Bilayer-DNA-Bilayer (BDB) structure. For three-layer formation, each layer must be bound by interactions between the DNA hydrogel and lipid membranes.
In addition, our other goal is to form the nuclear membrane-like pores by adding DNA origami structures in the gel layer. We designed spiral-shaped DNA origami structures, inducing the tubulation of inner membrane and membrane fusion between inner and outer membrane layers of BGB structure(Fig.1.b).
A detailed explanation of the methods for efficient BGB structure formation and of the DNA origami design for the pore formation is provided in the following sections.
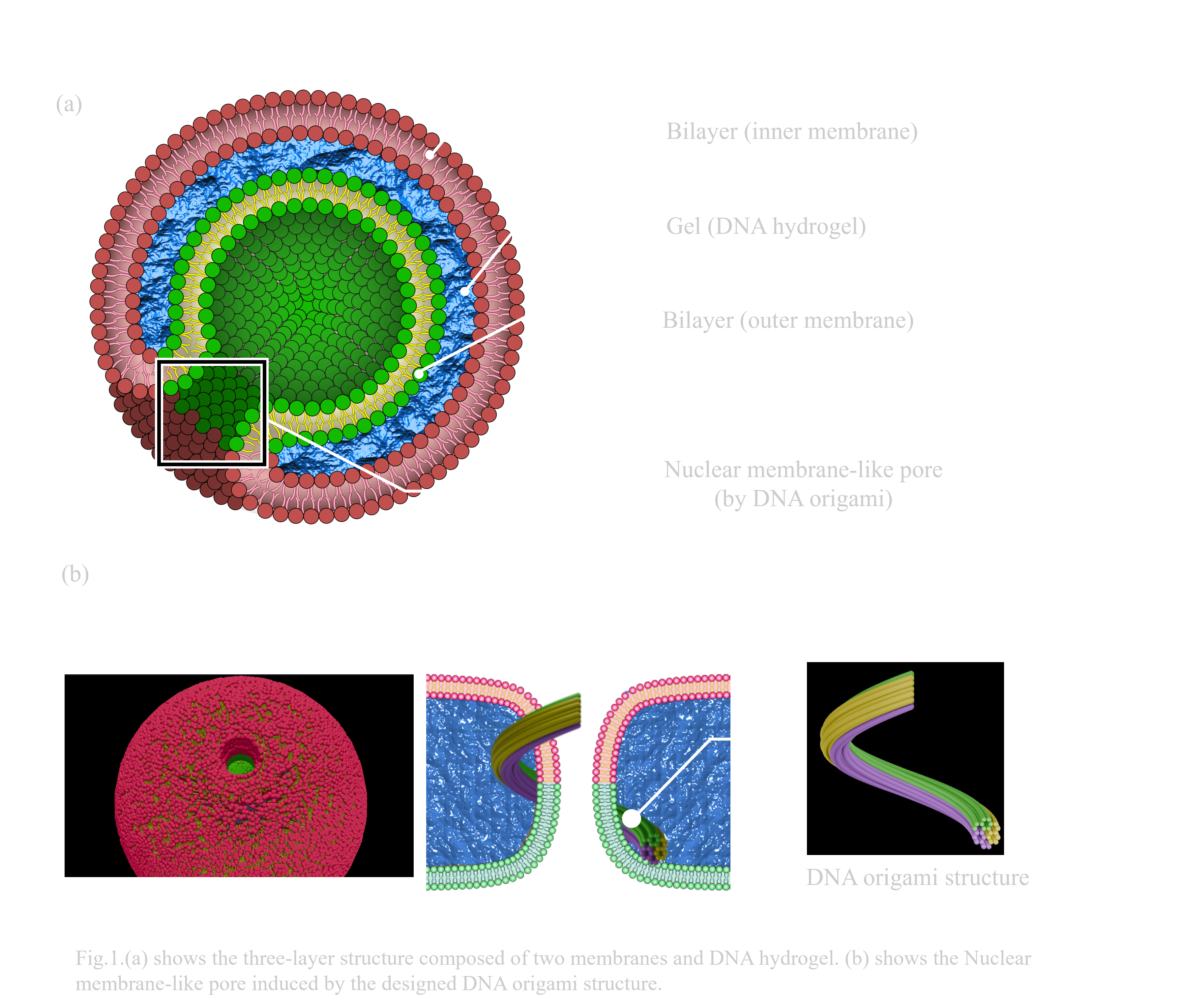
To form the BGB structure
BGB structures are formed in the order of Bilayer-DNA hydrogel-Bilayer from the innermost to the outermost layer by electrostatic interactions between liposome membranes and DNA strands. BGB structures are constructed by repeating the centrifugal sedimentation method. To achieve the BGB formation, we must consider the following three aspects:
- sufficient interaction between liposome membranes and DNA strands
- methods to confirm gel formation on liposomes
- the methods for verification of the BGB structure
The following sections detail the considerations for BGB structure formation in this order.
①Enough interactions between liposomes and DNA
a) Electrostatic interaction between membrane and DNA
To bind liposome membranes and DNA strands through electrostatic interaction,30 mol% of DOTAP were added to lipid membranes to obtain positive charge. Additionally, the ionic concentrations in the inner and outer solutions were adjusted to 50 mM KCl and 15 mM MgCl2 to enhance the electrostatic interaction. In the experiment to observe DNA localization on the lipid membrane, Rhodamine-PE (Rhod-PE) and AMCA-DNA were added to the lipid membrane and outer solution, respectively, allowing for the observation of the membrane and DNA positions.
b) To enhance the interaction
To enhance the interaction between the membrane and DNA, centrifugal sedimentation was optimized for the formation of the outermost membrane. This method consists of two elements: micelle formation using 100 mol% DOTAP and formation of the outer leaflet of the second membrane using subsequently added phospholipids. As the first step, 15 µl of BG solution is added to 100 mol% DOTAP (4.5 mM in mineral oil) and vortexed to form an emulsion. This step maximizes the interaction between the DNA hydrogel and the outer membrane. After forming an emulsion, lipids with a 4:3 molar ratio of DOPC (13.5 mM) are added to the emulsion to form the outer leaflet of the second membrane. This process contributes to stabilizing the outer membrane and prevents aggregation by reducing the charge of the outer leaflet.
②Formation and validation of DNA hydrogel
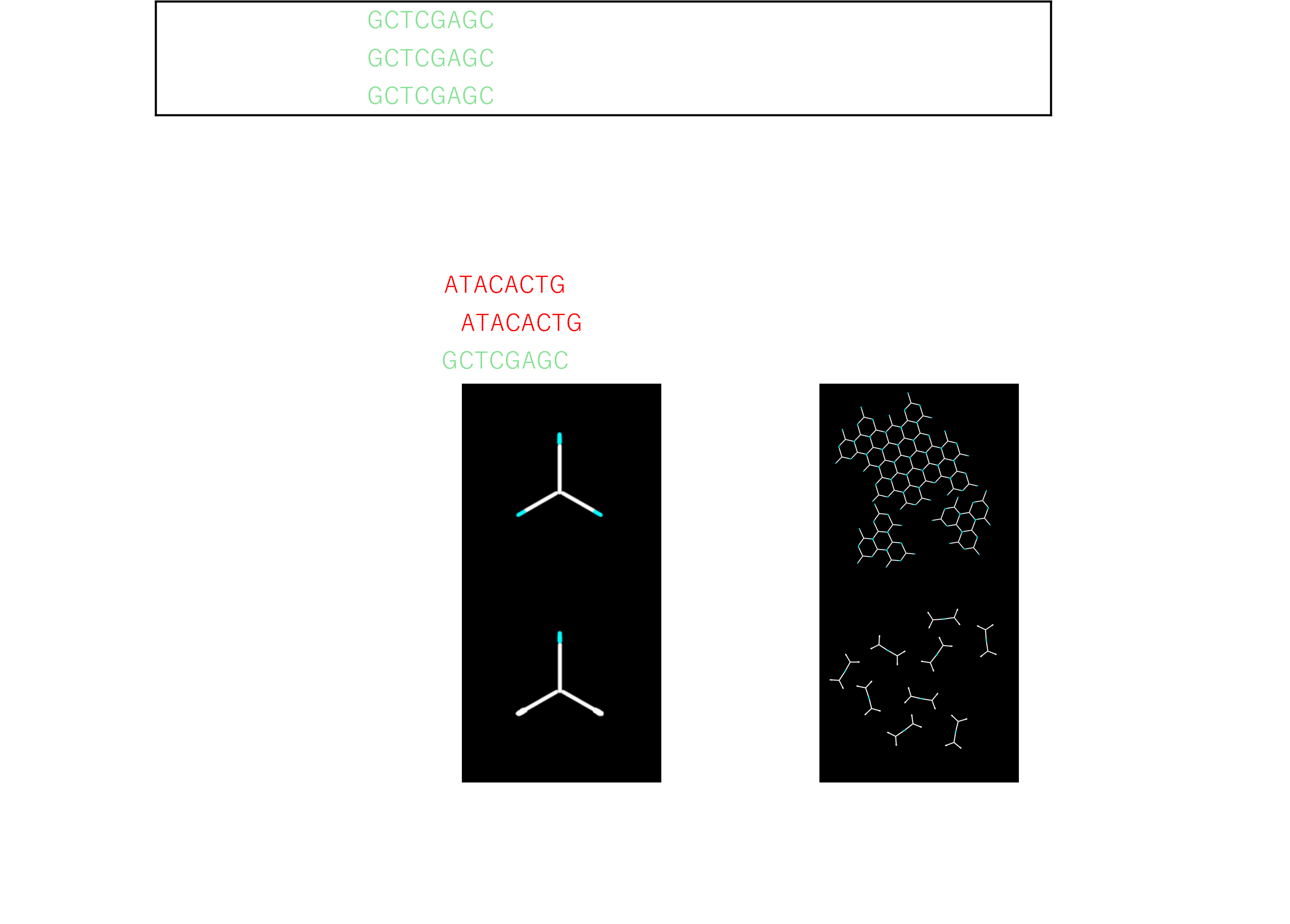
To form a DNA hydrogel network on liposomes, DNA Y-motifs with three sticky ends were designed. The total design was adopted from the previous study which formed DNA hydrogel on lipid membranes for liposome stabilization[1]. Each DNA strand has sticky ends with a palindromic sequence consisting of eight bases, facilitating binding between each DNA Y-motif structures. Y-motif DNA strands are categorized based on the presence or absence of sticky ends: G1, G2, and G3 have sticky ends on their 5’ ends, while S1, S2, and S3 do not. DNA strands with the same numbers have identical sequences except for the sticky ends (Fig. 2.a). Additionally, FAM-modified DNA strands (FAM-DNA) are designed with the same sequences of G3 DNA strands to observe the DNA hydrogel on the liposome membranes by confocal microscope observations. FAM-DNA is added at a 1:100 ratio relative to G3 DNA strands.
We can control the shape of DNA hydrogel networks by changing the combination of DNA strands. For instance, G1, G2, and G3 structures form DNA hydrogel networks by annealing from 90°C to 25°C at a rate of 1°C per 12 sec, while G1, G2, and S3 structures pair to form dimer structures (Fig.2. a).
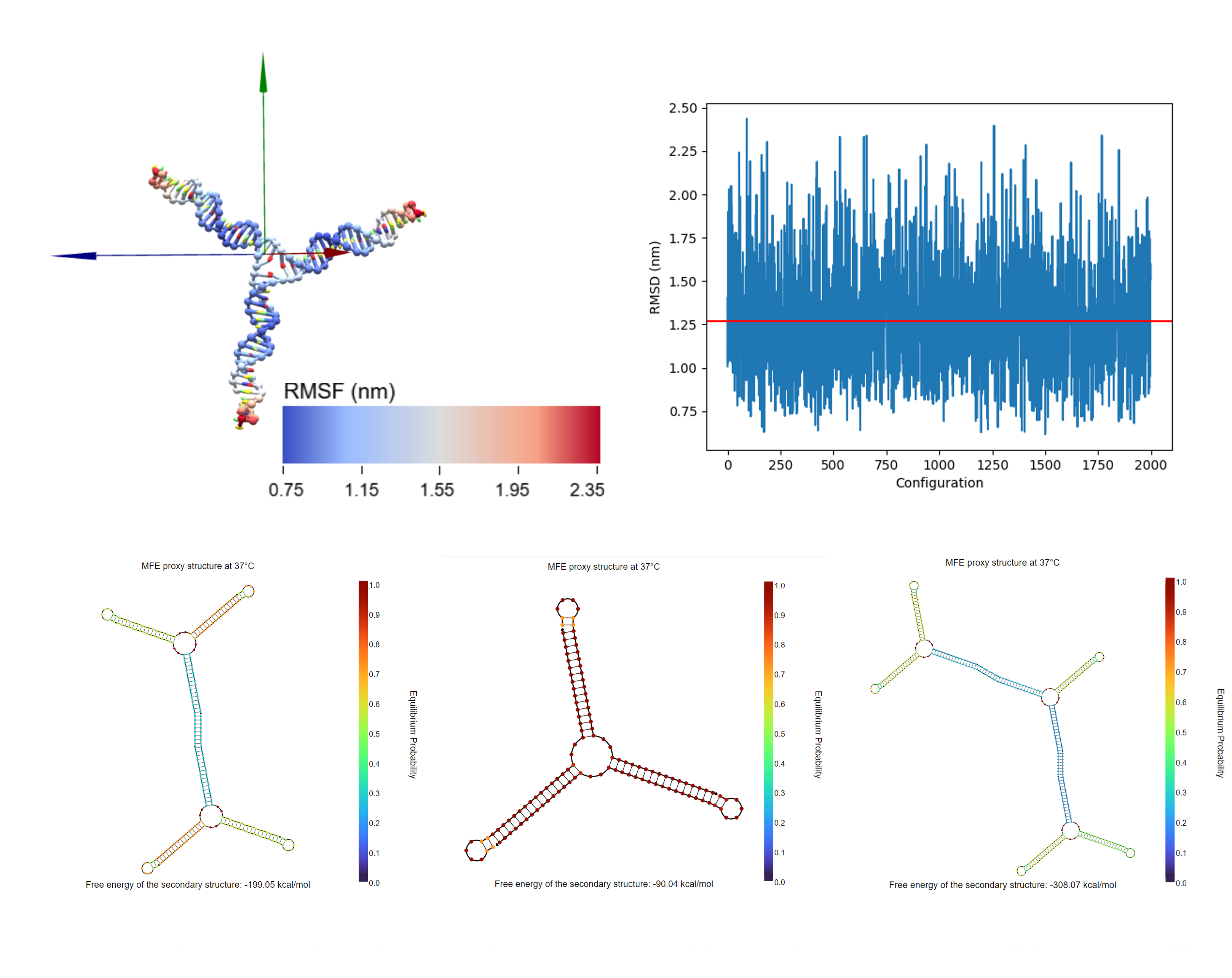
DNA hydrogel structures were designed and simulated using oxdna, showing that the average of RMSD scores were 1.25 nm. The simulation result revealed enough matches with the designed shape[2] (Fig. 2.b). Additionally, the hybridization between two or more Y-motif structures was confirmed by NUPACK(Fig.2.c).
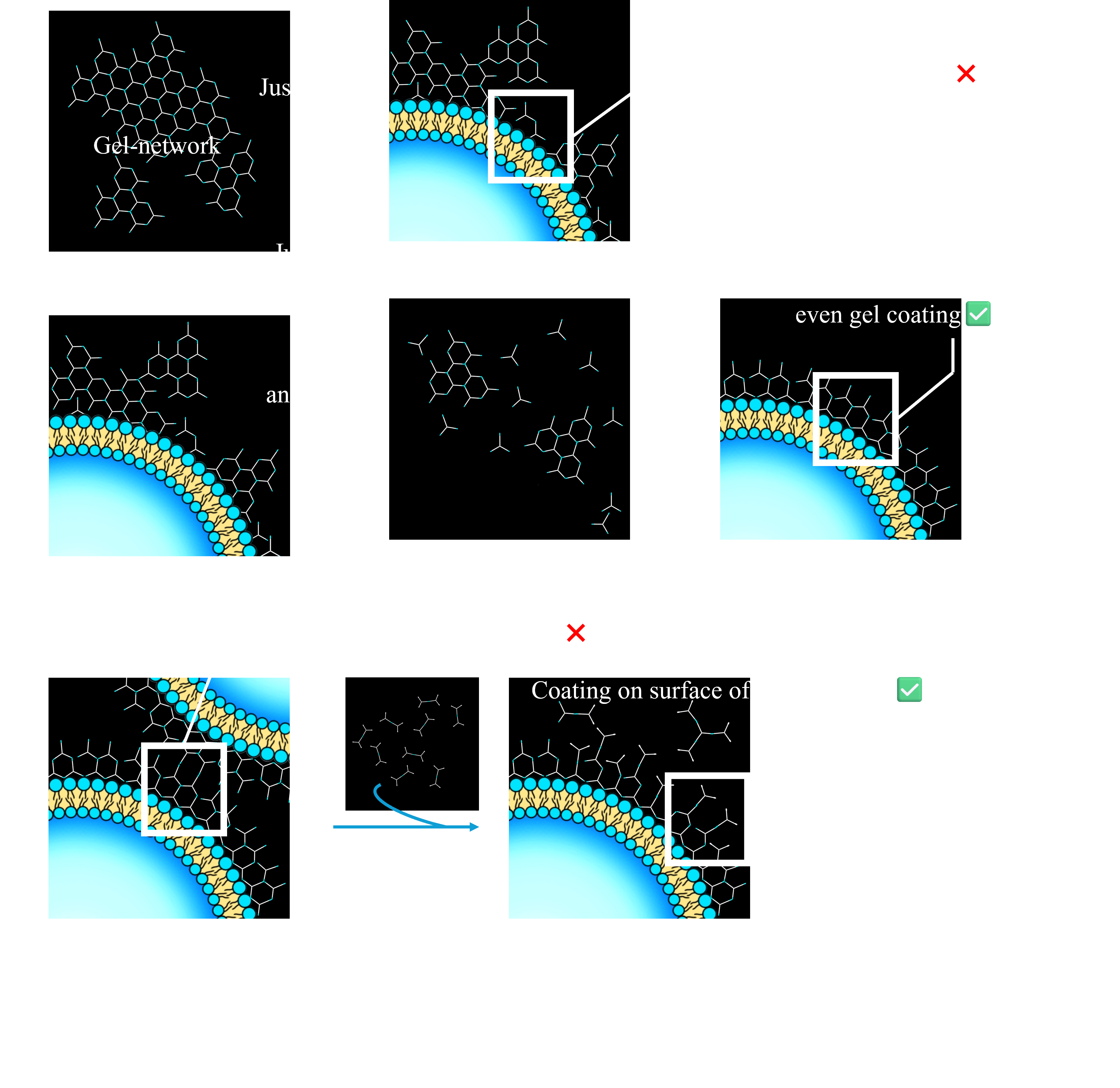
・To form DNA hydrogel evenly on the membrane surface
To form a uniform gel network on liposomes, there are several considerations different from gel formation in PCR tubes. One concern is that uneven gel formation may inhibit electrostatic interactions with the membrane(Fig.4.a). Additionally, exposure of sticky ends on the outer side of the BG structure can cause the aggregation of BG structures due to hybridization between exposed sticky ends. To address these issues, we developed a ‘Gel-Coating Method’.
In the first step of this method, a DNA hydrogel composed of G1, G2, and G3 (G1-G2-G3 Y-motif) was added to the liposome solution at a final concentration of 10 µM, and annealed from 50°C to 25°C at a rate of -1°C per 12 sec. This process promotes rehybridization of the DNA hydrogel on the liposomes, allowing for uniform hydrogel formation(Fig.4.b).
After this process, the Y-motif structure composed of G1, G2, and S3 DNA strands (G1-G2-S3 Y-motif) was added to the solution at a concentration of 1 µM. This induces a rearrangement from the G1-G2-G3 Y-motif to the G1-G2-S3 Y-motif on the surface of the BG structure, preventing the aggregation of liposomes by reducing exposed sticky ends(Fig.4.c).
③Varidation on the BGB structure-formation
・The position of three kinds of fluorescence for observation of three-layer structures
To observe BGB structures, NBD-PE modified lipids, AMCA-DNA strands, and Rhod-PE modified lipids were added in the inner membrane layer, DNA hydrogel layer, and outer membrane layer, respectively. All fluorescence signals are expected to be overlapped in desired structures.
・FRAP experiments for confirmation of the gel formation in BGB structures
To validate the gel formation on lipid membrane, FRAP experiment is planned. In this experiment, 100% intensity of 405 nm laser, 25% of 488 nm laser, and 100% of 561 nm laser are irradiated at 10000 ms stimulation time to the BGB layers. After photobleaching the specific area of fluorescent molecules on each layer, we observe the recovery of fluorescence intensity. In the desired BGB structure, only the gel layer is expected not to show the recovery of the fluorescence intensity.
To form the nuclear-ike pores on BGB structure
To achieve the nuclear-like pore formation on BGB structure, there are three considerations:
- the design of DNA origami structure for inducing membrane-tubulations
- the preparation of the negative-charged liposomes and ion controlling for membrane fusions
- Validation of the pore formation
①DNA origami-design
To induce tubulation of the liposome membrane, the DNA origami structure was designed based on a conventional vesicle tubulation study[3]. This structure can attach to a membrane via Chol-modified DNA strands within the structure, lifting the membrane. Each edge of the structure contains multiple linker DNA strands, facilitating the polymerization of the structure.
In this project, we modified the conventional DNA origami structure to allow formation using M13 DNA scaffold strands. The design was conducted in Cadnano software and simulated using oxDNA[4] (Fig. 4a,c). The simulation reveals that almost all bases of the structure fall within an RMSF value range of 2 nm to 5 nm, indicating reasonable stability for a complex 3D structure (Fig. 4c).
The designed data is available. - Download here-
②Negative charged liposome-formation and ion controlling for the membrane fusion
Membrane fusion is accelerated by electrostatic interactions between two membranes[5]. To induce this interaction, negatively charged liposomes containing 30 mol% POPG were prepared.
The negatively charged membranes, however, can repel DNA strands, posing a challenge for both DNA origami attachment and gel-coating with Chol-modified DNA hydrogels. To avoid this issue, the ion concentration of the liposome solution was adjusted to control the electrostatic shielding effect. For example, the KCl concentration in the outer solution is increased to 300 mM to allow attachment of Chol-modified DNA strands to the membrane for BG structures with DNA origami-lifted tubes. In contrast, the KCl concentration is decreased to 50 mM through buffer exchange when BG structures are encapsulated within the outer membrane, promoting membrane fusion.
③Validation of the pore formation
Forming pores inducing the influx of molecules from outer solution. FAM molecules were added in the outer solution of BGB structures, and observed the influx of FAM molecules in 1 hour.
Reference
- Chikako Kurokawa, Kei Fujiwara, Masamune Morita, Ibuki Kawamata, Yui Kawagishi, Atsushi Sakai, Yoshihiro Murayama, Shin-ichiro M. Nomura, Satoshi Murata, Masahiro Takinoue, and Miho Yanagisawa, PNAS, 2017, 114(28), 7228-7233.
- "Dashboard". oxDNA.org. https://oxdna.org/, (November 15, 2024)
- Michael W. Grome, Dr. Zhao Zhang, Dr. Frédéric Pincet, Prof. Chenxiang Lin, Angew. Chem. Int. Ed., 2018, Volume 57, Issue 19, 5330-5334.
- Nick Conway and Shawn Douglas. "Home". cadnano. 2016. https://cadnano.org/, (November 15, 2024)
- Liang Chen, Siping Liang, Yu Chen, Minhao Wu and Yuanqing Zhang, ACS Applied Materials & Interfaces, 2020, 12, 1, 96-105.